Methods in Molecular Virology: A Detailed Overview
Molecular virology is a field of study that delves into the
molecular aspects of viruses, focusing on their structure, replication, and
interactions with host cells. Researchers employ a variety of sophisticated
methods to investigate the intricate molecular mechanisms underlying viral
infections. Here is a detailed overview of some key methods used in molecular
virology:
- Polymerase
Chain Reaction (PCR):
- Purpose:
Amplification of viral nucleic acids.
- Description:
Polymerase Chain Reaction, commonly known as PCR, is a revolutionary
molecular biology technique that allows the amplification of specific DNA
sequences. Developed by Kary Mullis in 1983, PCR has become a cornerstone
in various scientific disciplines, including molecular virology.
Key Components:
- DNA
Template: The target DNA sequence to be amplified.
- Primers:
Short DNA sequences that flank the target region, serving as starting
points for DNA synthesis.
- DNA
Polymerase: Enzyme responsible for synthesizing a complementary DNA
strand based on the template.
Process:
- Denaturation:
The DNA template is heated to a high temperature (typically around
94–98°C), causing the DNA strands to separate, or denature, into single
strands.
- Annealing:
The reaction temperature is lowered (typically around 50–65°C), allowing
primers to bind (anneal) to their complementary sequences on the
single-stranded DNA template.
- Extension:
DNA polymerase synthesizes a new DNA strand by extending from the
primers. This occurs at a temperature optimal for the chosen DNA
polymerase (usually around 72°C).
- Amplification
Cycles: Steps 1-3 are repeated in cycles, doubling the amount of DNA
in each cycle. The number of cycles determines the final amount of the
amplified DNA.
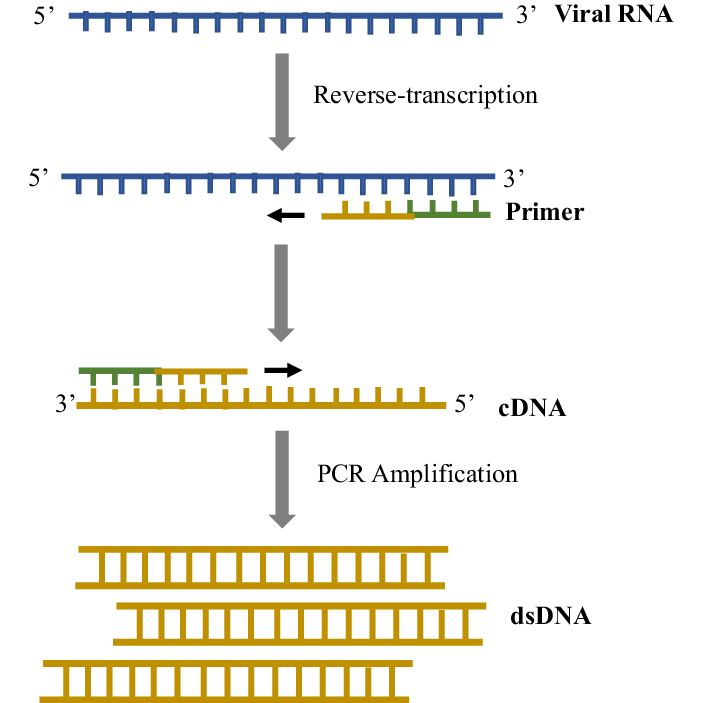
- Reverse
Transcription Polymerase Chain Reaction (RT-PCR):
- Purpose:
Amplification of viral RNA.
- Description:
Reverse Transcription Polymerase Chain Reaction, commonly known as
RT-PCR, is a molecular biology technique that combines reverse
transcription of RNA into complementary DNA (cDNA) with the subsequent
amplification of specific DNA sequences. RT-PCR is particularly crucial
for studying RNA viruses and analyzing gene expression.
Key Components:
- RNA
Template: The target RNA sequence to be converted into complementary
DNA.
- Reverse
Transcriptase: Enzyme responsible for synthesizing a complementary
DNA strand based on the RNA template.
- Primers:
Short DNA sequences that flank the target region, serving as starting
points for DNA synthesis.
- DNA
Polymerase: Enzyme responsible for amplifying the cDNA.
Process:
- Reverse
Transcription (RT): The RNA template is mixed with primers and
reverse transcriptase. Reverse transcriptase synthesizes a complementary
DNA strand from the RNA template, resulting in a cDNA molecule.
- Denaturation:
The cDNA is then subjected to denaturation, where it is heated to a high
temperature (typically around 94–98°C), causing the cDNA strands to
separate.
- Annealing:
Primers specific to the cDNA sequence bind to their complementary
sequences, serving as starting points for DNA synthesis.
- Extension:
DNA polymerase synthesizes a new DNA strand by extending from the
primers. This occurs at a temperature optimal for the chosen DNA
polymerase (usually around 72°C).
- Amplification
Cycles: Steps 2-4 are repeated in cycles, leading to the exponential
amplification of the cDNA.
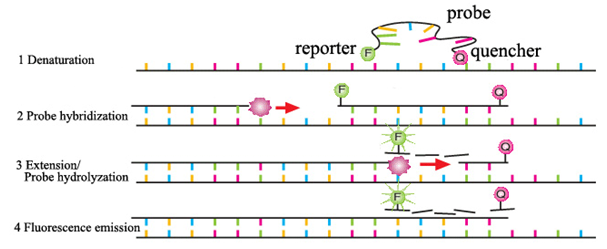
- Quantitative
PCR (qPCR):
- Purpose:
Accurate quantification of viral nucleic acids.
- Description:
Quantitative Polymerase Chain Reaction, commonly known as qPCR, is a
molecular biology technique that allows the real-time quantification of
DNA during the amplification process. It is an extension of traditional
PCR, providing a means to accurately measure and monitor the amount of
DNA in a sample.
Key Components:
- DNA
Template: The target DNA sequence to be amplified.
- Primers:
Short DNA sequences that flank the target region, serving as starting
points for DNA synthesis.
- DNA
Polymerase: Enzyme responsible for synthesizing a complementary DNA
strand based on the template.
- Fluorescent
Probes or DNA-binding Dyes: These are used to monitor the DNA
amplification in real-time.
Process:
- Denaturation:
The DNA template is heated to a high temperature (typically around
94–98°C), causing the DNA strands to separate.
- Annealing:
Primers bind to their complementary sequences on the single-stranded DNA
template. This occurs at a lower temperature (typically around 50–65°C).
- Extension:
DNA polymerase synthesizes a new DNA strand by extending from the
primers. This occurs at an optimal temperature for the chosen DNA
polymerase (usually around 72°C).
- Fluorescence
Detection: As DNA is synthesized, fluorescent probes or DNA-binding
dyes emit fluorescence. The increase in fluorescence is proportional to
the amount of DNA being synthesized.
- Real-Time Monitoring: The fluorescence is monitored after each amplification cycle in real-time, allowing for the continuous quantification of the DNA during the PCR process.
- Northern
Blotting:
- Purpose:
Detection of viral RNA.
- Description:
Northern blotting is a molecular biology technique used to study and
analyze RNA molecules. It allows researchers to detect and characterize
specific RNA transcripts based on size and abundance.
Key Components:
- RNA
Sample: The target RNA molecules to be analyzed.
- Gel
Electrophoresis System: Used to separate RNA molecules based on size.
- Membrane
(typically nylon or nitrocellulose): Transfers separated RNA from the
gel for further analysis.
- RNA
Probes: Radioactively or chemically labeled single-stranded DNA or
RNA sequences that are complementary to the target RNA.
Process:
- Gel
Electrophoresis: RNA samples are separated by size through gel
electrophoresis. The gel is typically made of agarose or polyacrylamide.
- Transfer
to Membrane: The separated RNA molecules are transferred from the gel
to a membrane through a process called blotting. This preserves the
spatial arrangement of the RNA bands on the gel.
- Hybridization:
The membrane is incubated with a labeled RNA probe that is complementary
to the target RNA sequence. The probe binds specifically to the target
RNA.
- Washing:
Excess and nonspecifically bound probes are washed away, leaving only the
specifically bound probes on the membrane.
- Detection:
The membrane is exposed to X-ray film or a phosphorimager to visualize
the labeled RNA bands. The intensity of the bands corresponds to the
abundance of the target RNA molecules.
- Western
Blotting:
- Purpose:
Detection of viral proteins.
- Description:
Western blotting is a widely used molecular biology technique that allows
the detection and analysis of specific proteins within a complex mixture.
It involves the separation of proteins based on size, followed by their
transfer to a membrane and subsequent detection using specific
antibodies.
Key Components:
- Protein
Sample: The complex mixture containing the proteins of interest.
- Polyacrylamide
Gel Electrophoresis (SDS-PAGE): Used to separate proteins based on
size.
- Transfer
System: Transfers separated proteins from the gel to a membrane.
- Primary
Antibodies: Specific antibodies that bind to the target protein.
- Secondary
Antibodies: Conjugated to enzymes or fluorescent tags to detect bound
primary antibodies.
- Detection
System: Chemiluminescent substrates or fluorescent detection methods.
Process:
- Protein
Separation: Proteins are separated by size using SDS-PAGE. The gel is
then exposed to an electric field, causing the proteins to migrate based
on their molecular weight.
- Transfer
to Membrane: Proteins are transferred from the gel to a membrane,
typically made of nitrocellulose or PVDF. This preserves the spatial
arrangement of the proteins on the gel.
- Blocking:
The membrane is treated with a blocking agent to prevent nonspecific
binding of antibodies.
- Incubation
with Primary Antibodies: The membrane is incubated with specific
primary antibodies that bind to the target protein.
- Washing:
Excess and nonspecifically bound antibodies are washed away.
- Incubation
with Secondary Antibodies: The membrane is then incubated with
secondary antibodies that recognize and bind to the primary antibodies.
The secondary antibodies are often conjugated to enzymes or fluorescent
tags.
- Detection:
The presence of the target protein is visualized by adding a substrate
for the enzyme (in the case of enzyme-conjugated antibodies) or by using
a fluorescence imaging system. This produces a signal that can be
captured and quantified.
- Viral
Genome Sequencing:
- Purpose:
Determining the nucleotide sequence of viral genomes.
- Description:
Viral genome sequencing is a molecular biology technique that involves
determining the complete nucleotide sequence of a viral genome. This
process provides crucial information about the genetic makeup, structure,
and evolution of viruses.
Key Components:
- Viral
Sample: The source material containing the viral genetic material
(RNA or DNA).
- Nucleic
Acid Extraction: The isolation of viral RNA or DNA from the sample.
- Library
Preparation: The creation of a library of DNA fragments for
sequencing.
- Sequencing
Technology: High-throughput sequencing platforms that determine the
sequence of nucleotides.
- Bioinformatics
Analysis: Computational tools used to assemble and analyze the
sequence data.
Process:
- Nucleic
Acid Extraction: Viral RNA or DNA is extracted from the sample using
specialized techniques, separating it from cellular components.
- Library
Preparation: The extracted genetic material is converted into a
library of DNA fragments. This step often involves DNA fragmentation,
adapter ligation, and PCR amplification.
- Sequencing:
The prepared library is loaded onto a high-throughput sequencing
platform, such as next-generation sequencers. These machines read the
sequence of nucleotides in the DNA fragments.
- Assembly:
Bioinformatics tools are employed to assemble the sequenced fragments
into a complete viral genome. This step involves aligning and overlapping
the short DNA reads to reconstruct the full-length viral genome.
- Annotation:
The identified viral genes and other features are annotated to provide
information about open reading frames, regulatory elements, and potential
functional proteins.
- Analysis:
Comparative genomics, phylogenetic analysis, and other bioinformatics
tools are used to study the genetic variations, relatedness to other
viruses, and potential implications for host interactions.
- Gene
Expression Profiling:
- Purpose:
Analyzing changes in host gene expression during viral infection.
- Description:
Gene expression profiling is a molecular biology technique that involves
the systematic analysis of the expression levels of genes in a given
biological sample. It provides insights into which genes are active and
to what extent, offering a comprehensive view of the transcriptional
activity within cells or tissues.
Key Components:
- RNA
Sample: The source material containing RNA, often extracted from
cells, tissues, or organisms.
- mRNA
Isolation: The isolation of messenger RNA (mRNA), which represents
the actively transcribed genes.
- cDNA
Synthesis: Conversion of mRNA into complementary DNA (cDNA) using
reverse transcription.
- Microarray
or RNA Sequencing: Technologies used to measure the abundance of cDNA
corresponding to different genes.
- Bioinformatics
Analysis: Computational tools to interpret and analyze the
large-scale gene expression data.
Process:
- RNA
Extraction: Total RNA is extracted from the biological sample,
preserving the RNA's integrity.
- mRNA
Isolation: The isolated RNA is then enriched for mRNA, as mRNA
represents the actively transcribed genes.
- cDNA
Synthesis: Reverse transcription converts the isolated mRNA into
complementary DNA (cDNA). This step reflects the abundance of each mRNA
molecule.
- Labeling:
The cDNA is often labeled with fluorescent markers or other detectable
tags for identification.
- Microarray
Hybridization or RNA Sequencing: The labeled cDNA is applied to a
microarray (for microarray-based profiling) or subjected to RNA
sequencing (for next-generation sequencing-based profiling). These
technologies measure the abundance of different cDNA molecules,
representing various genes.
- Data
Analysis: Bioinformatics tools are employed to analyze the
large-scale gene expression data. This includes normalization,
statistical analysis, and interpretation of gene expression patterns.
- Visualization:
The results are visualized in the form of heatmaps, gene expression
profiles, or other graphical representations, highlighting upregulated
and downregulated genes.
These methods collectively form the backbone of molecular
virology, allowing researchers to unravel the complexities of viral infections
at the molecular level. Advancements in these techniques contribute
significantly to our understanding of viral pathogenesis and aid in the
development of novel antiviral strategies.
Frequently Asked Questions (FAQs) about Molecular
Virology:
- What
is molecular virology?
- Molecular
virology is a branch of virology that focuses on the study of viruses at
the molecular level. It investigates the structure, replication, and
interactions of viruses with host cells, using molecular biology
techniques to understand the underlying mechanisms of viral infections.
- Why
is studying molecular virology important?
- Understanding
molecular virology is crucial for developing effective antiviral
therapies, vaccines, and diagnostic tools. It provides insights into the
genetic makeup of viruses, their replication strategies, and host
responses, aiding in the prevention and treatment of viral diseases.
- What
are the key methods used in molecular virology?
- Key
methods in molecular virology include PCR, RT-PCR, qPCR for nucleic acid
amplification, Northern and Western blotting for RNA and protein
detection, viral genome sequencing, gene expression profiling,
protein-protein interaction studies, CRISPR/Cas9 for gene editing, and
fluorescence microscopy for real-time visualization.
- How
does PCR contribute to molecular virology research?
- PCR
(Polymerase Chain Reaction) is essential in molecular virology for
amplifying specific DNA sequences, enabling the detection and
quantification of viral genomes. It plays a key role in studying viral
DNA and understanding the dynamics of viral replication.
- What
is the significance of gene expression profiling in molecular virology?
- Gene
expression profiling helps researchers analyze changes in host gene
expression during viral infections. This method provides insights into
how viruses modulate cellular processes and elicit host responses, aiding
in the identification of potential therapeutic targets.
- How
does CRISPR/Cas9 technology contribute to virology research?
- CRISPR/Cas9
technology allows precise gene editing in viral genomes or host cells. In
virology research, it is used to study gene function, develop antiviral
strategies, and investigate the impact of genetic modifications on viral
replication.
- What
is the role of fluorescence microscopy in studying viral infections?
- Fluorescence
microscopy and live-cell imaging enable real-time visualization of viral
particles and infected cells. This method helps researchers observe the
dynamics of viral entry, replication, and assembly within host cells,
providing valuable insights into the spatial and temporal aspects of
viral infections.
- How
does viral genome sequencing contribute to our understanding of viruses?
- Viral
genome sequencing, using next-generation sequencing technologies, allows
the complete analysis of viral genomes. This method is crucial for
understanding viral diversity, evolution, and identifying potential drug
targets.
- Why
study protein-protein interactions in molecular virology?
- Protein-protein
interaction studies are essential for understanding the molecular events
during viral infection. Techniques such as yeast two-hybrid assays or
co-immunoprecipitation help identify and characterize interactions
between viral and host proteins, revealing key pathways involved in viral
replication and pathogenesis.
- What
are the applications of molecular virology in medicine and public health?
- Molecular
virology has numerous applications in medicine and public health,
including the development of antiviral drugs, vaccines, and diagnostic
tools. It provides the foundation for understanding viral diseases and
developing strategies to control and prevent their spread.






0 Comments